Developmental Bifurcations
An individual animal is made up of many different kinds of cells, from neurons to gametes, and exhibits significant geometric asymmetry, in having, for example, a head at one end and feet at the other. Yet, all animals start as a single, mostly symmetric cell: a fertilized egg that has the functional potential to produce any cell type (pluripotent) and to induce the shape changes necessary to yield its matured, complex geometry. The past century of developmental biology research has distinguished cell types and identified molecules that induce them, but the dynamic processes through which cells commit to specific fates and establish robust spatial patterns are not well understood.
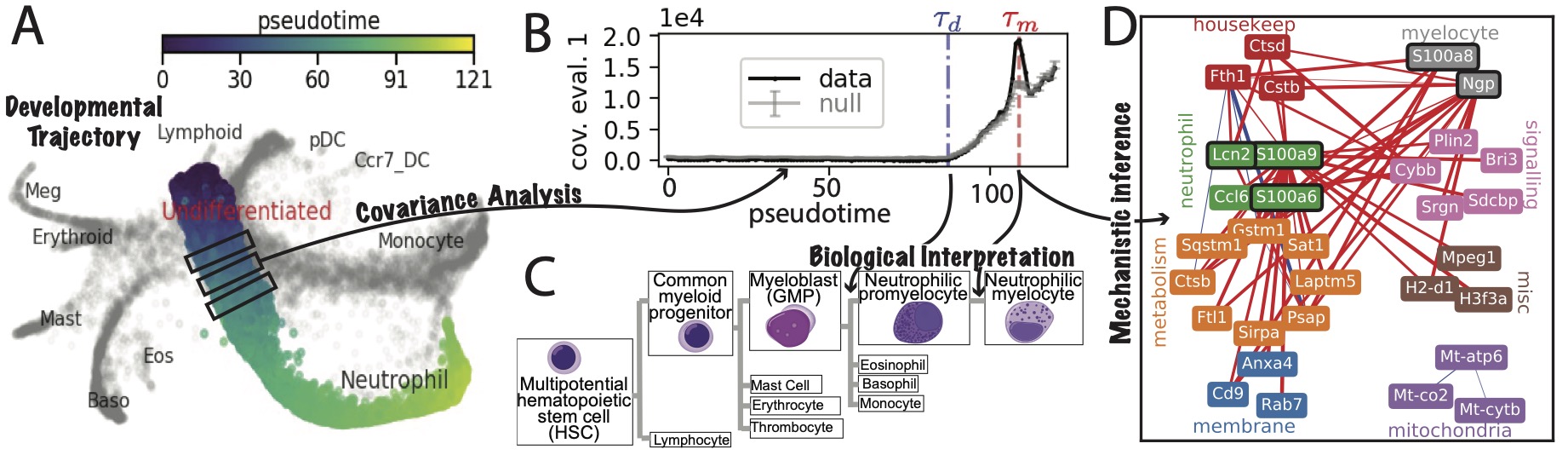
I am applying concepts from dynamical systems theory to big biological data to bridge the scales between cell fate and molecular mechanism. I have recently shown how bifurcations – the signature of fundamental change in classical, physical dynamical systems, like a beam buckling, or a power-line network crashing- can be detected during development from single-cell RNA-seq data. Moreover, information about the underlying dynamical system guiding the transition, which is normally hidden, because it operates at a faster timescale than we can measure, is inferrable from the data. I am currently working to push this research further from multiple angles, by studying bifurcations during morphogenesis and using simple models to determine if we can predict the results of developmental perturbation experiments directly from gene expression data.